 Computational Biophysics
Computational Biophysics
and Materials Science Group
-Department of Materials Science and Engineering
My long-term research goal is to explore molecular mechanisms of cellular systems and materials as well as to develop new computational tools in order to understand fundamental physical principles behind biological and materials phenomena.
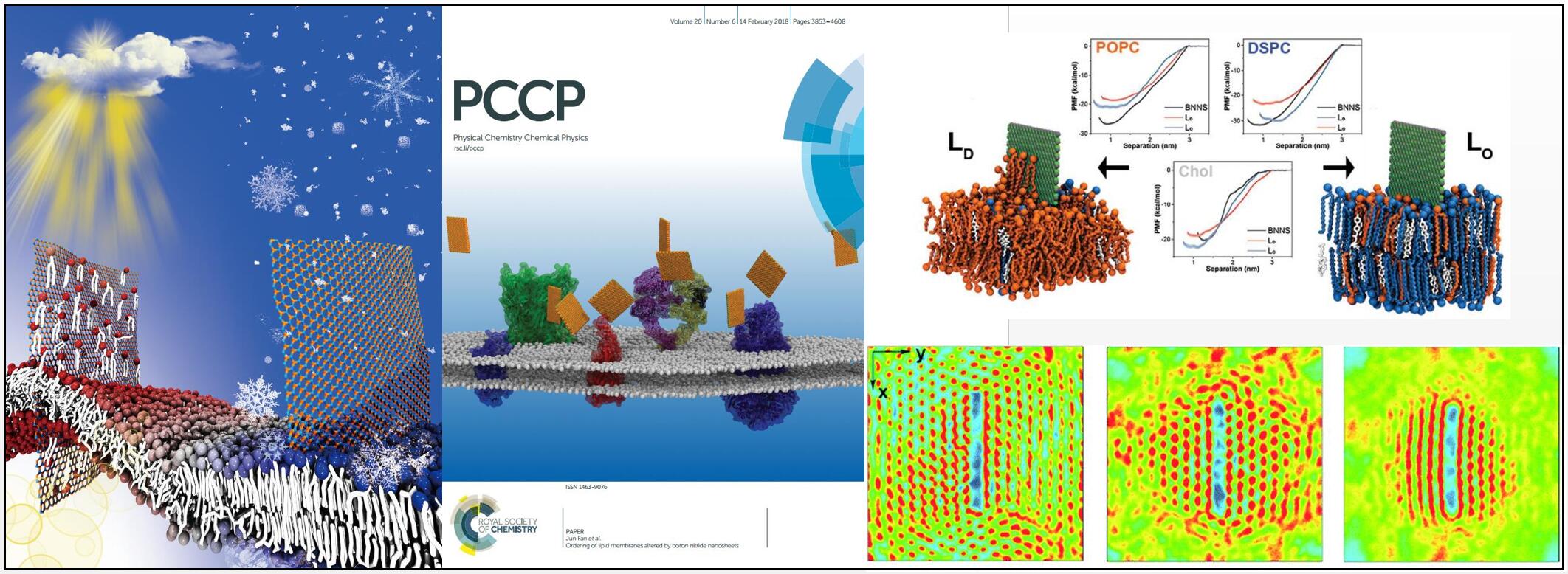
As the rapid development of nanomaterials and their emerging biomedical and biological applications such as drug/gene delivery and nanosensing, the interaction between nanomaterials and biological entities plays an increasingly important role in nanoscience. In particular, bio-nano interactions could interfere functions of proteins, lipids, etc., and even damage the structural integrity of cells. Thus, the cytotoxicity of nanomaterials has caused serious concerns over the safety and sustainability of nanotechnology. On the other hand, the toxicity of nanomaterials can also be utilized in antibacterial and antifouling applications. We perform MD simulations to study the interaction between nanoparticles/nanotubes/nanosheets and lipid membranes/proteins/DNA/organelles, providing insights into physics in bio-nano systems.
We have studied the interaction between 2D nanomaterials (nanosheets) and lipid membranes recently. Hydrophobic nanosheets could extract lipids and then insert into lipid membranes, which is very important for the cytotoxicity of nanosheets. Interestingly, we found that the lipid extraction behavior depends on the phase state of lipids that can be changed by temperature, lipid components, etc. In addition, the insertion of nanosheets into lipid membranes could lead to the ordering of lipids, which may perturb the function of transmembrane proteins.
References
- Yonghui Zhang, Chun Chan, Zhen Li, Jiale Ma, Qiangqiang Meng, Chunyi Zhi, Hongyan Sun, and Jun Fan*. Nanotoxicity of Boron Nitride Nanosheet to Bacterial Membranes. Langmuir, 18: 6179 (2019), Selected Supplementary Cover Story.
- Zhen Li, Yonghui Zhang, Jiale Ma, Qiangqiang Meng, and Jun Fan*. Modelling Interactions between Liposomes and Hydrophobic Nanosheets. Small, 15: 1804992 (2019). Inside Back Cover Story.
- Zhen Li, Yonghui Zhang, Chun Chan, Chunyi Zhi*, Xiaolin Cheng*, and Jun Fan*. Temperature-Dependent Lipid Extraction from Membranes by Boron Nitride Nanosheets. ACS Nano, 12: 2764-2772 (2018).
- Yonghui Zhang, Chun Chan, Zhen Li, Jia Le Ma, Qiangqiang Meng, Xiaolin Cheng, and Jun Fan*. Lipid Extraction by Boron Nitride Nanosheets from Liquid-ordered and Liquid-disordered Nanodomains. Nanoscale, 10: 14073 (2018).
- Yonghui Zhang, Zhen Li, Chun Chan, Jiale Ma, Chunyi Zhi, Xiaolin Cheng, and Jun Fan*. Ordering of Lipid Membranes Altered by Boron Nitride Nanosheet. Phys. Chem. Chem. Phys., 20: 3903-3910 (2018), Inside Cover Story.
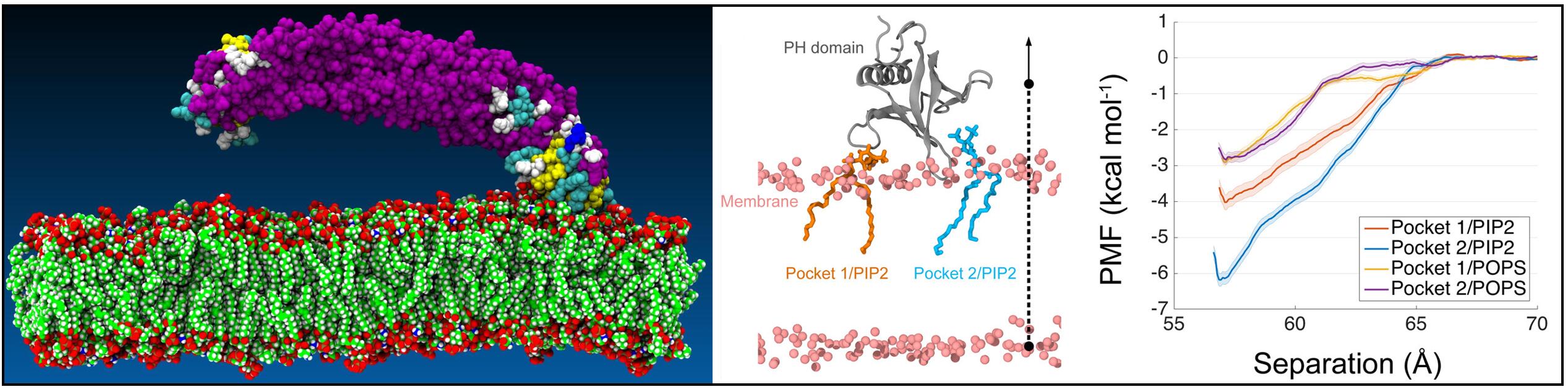
Peripheral membrane proteins and transmembrane proteins could influence the physical state and functions of lipids, and vice versa. Utilizing molecular dynamics (MD) simulations and other computational analyses, we aim to explore the relationship or coupling between proteins and lipids, including the membrane remodeling by BAR (Bin-Amphiphysin-Rvs) family proteins, the reversible gating of protein channels stimulated by the lateral pressure of lipid membranes, etc.
The BAR (Bin-Amphiphysin-Rvs) domain undergoes dimerization to produce a curved protein structure, which superimposes onto membrane through electrostatic interactions to sense and impart membrane curvature. In some cases, a BAR domain also possesses an amphipathic helix that inserts into the membrane to induce curvature. ACAP1 (Arfgap with Coil coil, Ankyrin repeat, and PH domain protein 1) contains a BAR domain. Here, we show that this BAR domain can neither bind membrane nor impart curvature, but instead requires a neighboring PH (Pleckstrin Homology) domain to achieve these functions. Specific residues within the PH domain are responsible for both membrane binding and curvature generation. The BAR domain adjacent to the PH domain instead interacts with the BAR domains of neighboring ACAP1 proteins to enable clustering at the membrane. Thus, we have uncovered the molecular basis for an unexpected and unconventional collaboration between PH and BAR domains in membrane bending.
References
- Chun Chan, Xiaoyun Pang, Yan Zhang, Tongxin Niu, Shengjiang Yang, Daohui Zhao, Jian Li, Lanyuan Lu, Victor W. Hsu, Jian Zhou*, Fei Sun*, and Jun Fan*. ACAP1 Assembles into an Unusual Protein Lattice for Membrane Deformation through Multiple Stages.Plos Computational Biology, 15(7): e1007081 (2019).
- Seung-Yeol Park, Jia-Shu Yang, Zhen Li, Xiaohong Zhu, Maria Ericsson, Andrew J. Morris, Jun Fan, and Victor W. Hsu. The Late Stage of COPI Vesicle Fission Requires Shorter Forms of Phosphatidic Acid and Diacylglycerol. Nat.Comm., 10: 3409 (2019).
- Chun Chan, Lanyuan Lu, Fei Sun, and Jun Fan*. Molecular Details of the PH Domain of ACAP1BAR-PH Protein Binding to PIP-Containing Membrane. J. Phys. Chem. B, 121: 3586-3596 (2017).
- Chun Chan, Haohua Wen, Lanyuan Lu, and Jun Fan*, Multiscale Molecular Dynamics Simulations Membrane Remodelling by BAR Family Proteins. Chinese Physics B, 25: 018707 (2016), Invited Review.
- Xiaoyun Pang, Jun Fan, Yan Zhang, Kai Zhang, Bingquan Gao, Jun Ma, Jian Li, Yuchen Deng, Qiangjun Zhou, Edward H. Egelman, Victor W. Hsu*, and Fei Sun*. A PH Domain in ACAP1 Possesses Key Features of the BAR Domain in Promoting Membrane Curvature. Developmental Cell, 31: 73 (2014). Highlighted in A Novel Twist In Membrane dePHormation Developmental Cell, 31: 3 (2014).
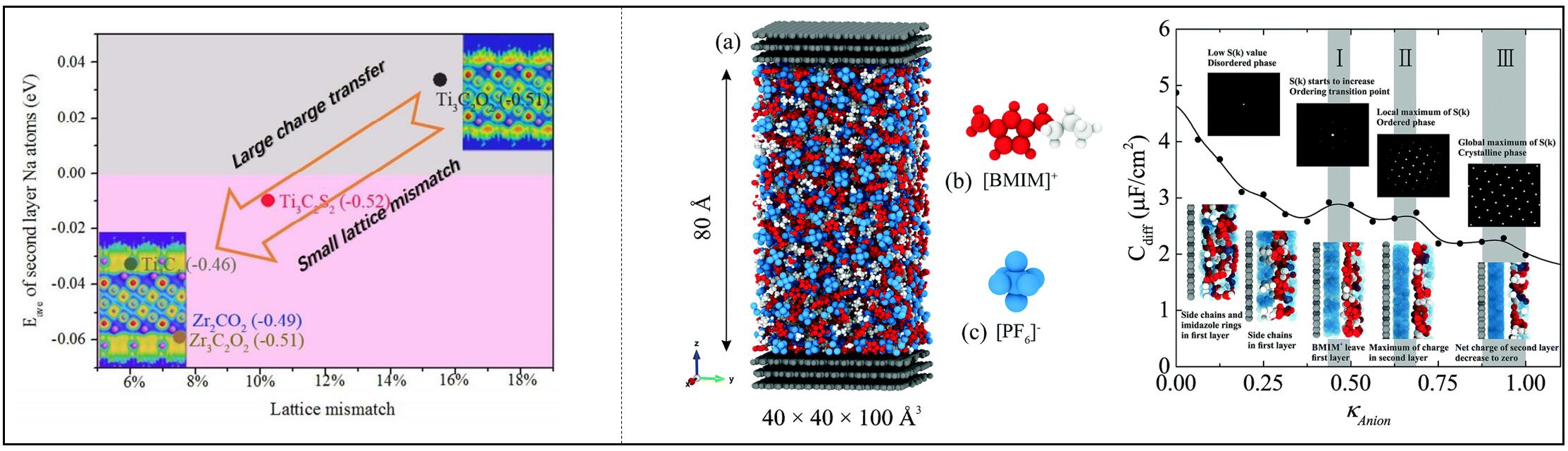
Combining DFT calculations and MD simulations, we explore physics in energy-storage materials such as Li-ion batteries, Na-ion betteries, and supercapacitors. With DFT calculations, we evaluate the energy-storage properties of nanomaterials (especially 2D materials), and predict novel electrode materials. MD simulations are implemented to explore the structural characteristics and physical behaviors of ions and electrolytes on the interface between electrodes and electrolytes.
We have studied the storage of Na-ion on MXene-family electrodes, and concluded that large charge transfer and small lattice mismatch are beneficial to the storage of Na-ions. We investigated the structure of ionic liquid electrolytes in electric double layers of supercapacitors, and clarified the charge-driven ordering transition and the role of electrolyte structures.
References
- Na Li, Qiangqiang Meng, Xiaohong Zhu, Zhen Li, Jiale Ma, Changxiong Huang, Jun Song and Jun Fan*. Lattice constants-dependent Anchoring Effect of MXenes for Lithium-sulfur (Li-S) Batteries: A DFT Study. Nanoscale, 11: 8485 (2019).
- Jiale Ma, Qiangqiang Meng, Chun Chan, Zhen Li, Yonghui Zhang, and Jun Fan*. Alkyl Tail Aggregations Break Long Range Ordering of Ionic Liquids Confined in Sub-nanometer Pores. J. Phys. Chem. C, 122: 27314–27322 (2018).
- Jiale Ma, Qiangqiang Meng, and Jun Fan*. Charge Driven Lateral Structural Evolution of Ions in Electric Double Layer Capacitor Strongly Correlates with Differential Capacitance. Phys. Chem. Chem. Phys., 20: 9054-8063 (2018).
- Qiangqiang Meng, Jia Le Ma, Yonghui Zhang, Zhen Li, Alice Hu, Ji-Jung Kai, and Jun Fan*. Theoretical Investigation of Zirconium Carbide Mxenes as Prospective High Capacity Anode Materials for Na-ion Batteries. J. Mater. Chem. A, 6: 13652 (2018). This article is part of the themed collection: 2018 JMCA HOT Papers.
- Qiangqiang Meng, Jiale Ma, Alice Hu, Chunyi Zhi, and Jun Fan*. The S-functionalized Ti3C2 Mxene as a High Capacity Electrode Material for Na-ion Batteries: A DFT Study. Nanoscale, 10: 3385-3392 (2018).
- Qiangqiang Meng, Alice Hu, Chunyi Zhi, and Jun Fan*. Theoretical Prediction of MXene-like Structure Ti3C4 as a High Capacity Electrode Material for Na ion Batteries. Phys. Chem. Chem. Phys., 19: 29106-29113 (2017). This article is part of the themed collection: 2017 PCCP HOT Articles.
